by Florian Rämisch and Peter Mühleder on behalf of the diggr project
The diggr project released its swiss-army-knife toolset as open source software this week. Implemented in Python, the diggrtoolbox provides various helpful functions for researchers doing data analysis in the game and game culture studies. Often required methods and tools are packaged to be easily included in your existing code.
Platform name standardization
The library includes methods to acquire the latest diggr platform mapping which provides a unique set of standardized platform spellings, as well as mappings for various popular video game databases. See also this blogpost.
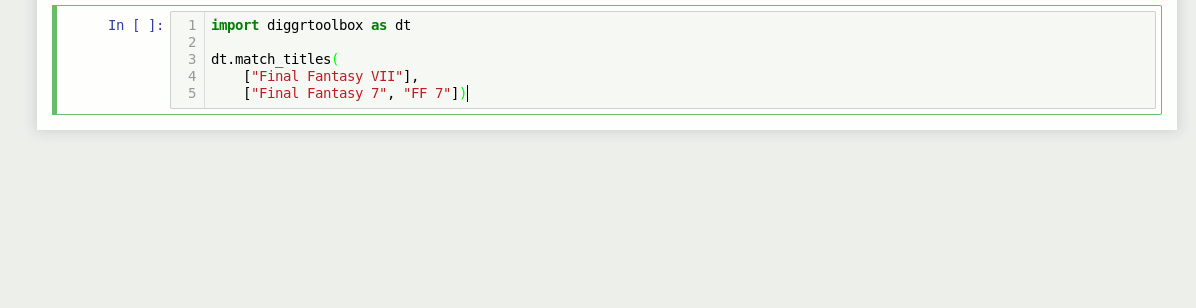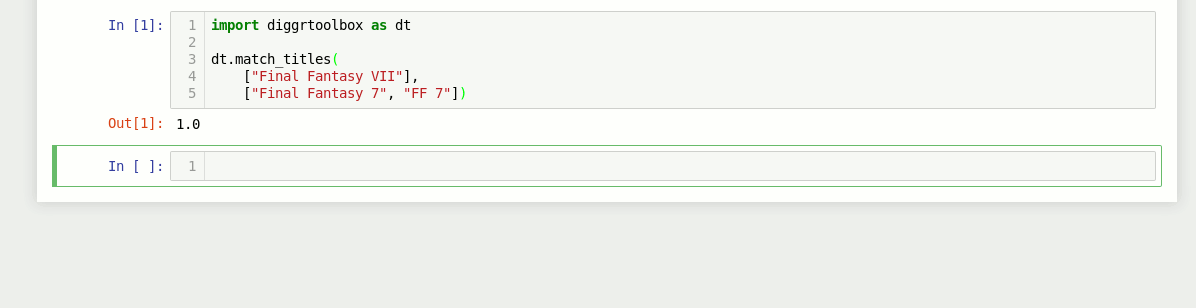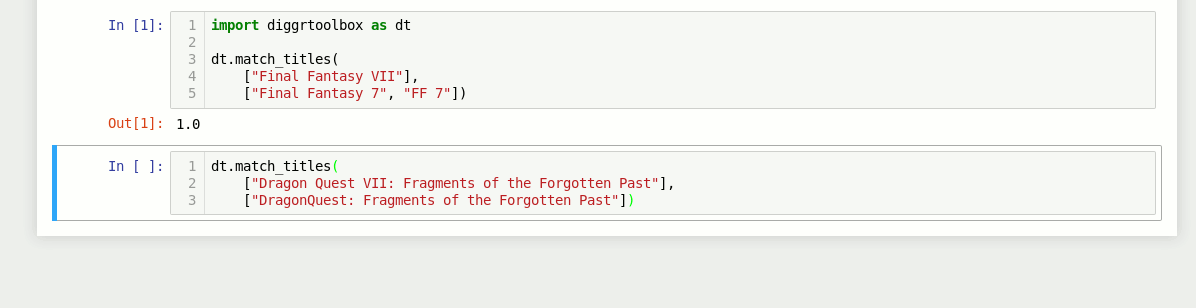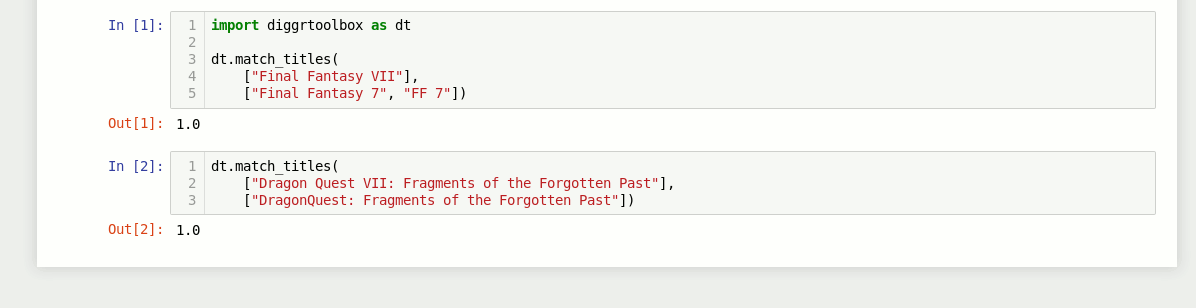
Game title matching algorithm
The library also contains an algorithm for matching video game titles. It returns a value between 0 and 1, based on the titles’ similarity. Numbers in game titles get detected and evaluated separately.
Further contents
Overall, the diggrtoolbox library contains the following subpackages:
- configgr: Provides a class which helps to keep a program wide configuration.
- deepget: A small helper easing access to data in deeply nested dicts/list, by separating the definition of the route and actual call.
- linking: Contains the matching algorithm for video game titles.
- platform_mapping: Methods to acquire the latest diggr platform mapping.
- schemaload: Verify JSON data loaded into a program is consistent with a given JSON-schema.
- standardize: String standardization tools, e.g. to get data ready for string matching etc. Helps removing most of the unwanted symbols, and characters complicating string comparison.
- treeexplore: Methods to compare deeply nested dict and list data structures. Especially helpful for searching for specific strings/values in deeply nested dicts.
- zipaccess: Allows access to a JSON document in a ZIP-File.
Acquisition, Usage, Documentation, Examples
The diggrtoolbox is open source, the source code is to be found on github. After cloning or downloading it can be installed with Pip.
The full documentation is to be found in the repository itself or online at Read The Docs and contains more information on all packages, modules and methods. Furthermore, we made great effort to provide a set of useful examples to demonstrate the possible applications of the library as well as integration in existing research environments.